what’s new
in the tidyverse
in 2023?
🔗 mine.quarto.pub/tidyverse-2023
💻 github.com/mine-cetinkaya-rundel/tidyverse-2023
duke university + posit
2023-05-30
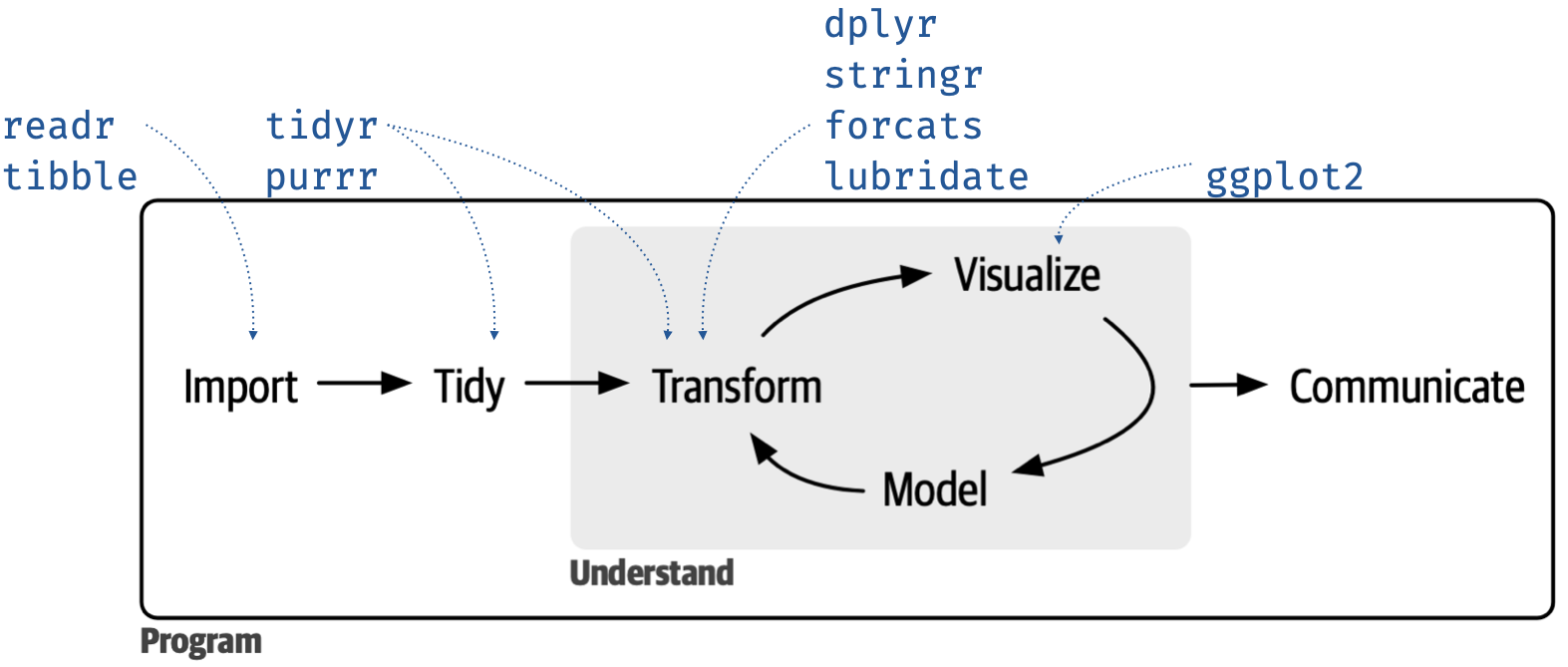
principles of the tidyverse
tidyverse
meta R package that loads nine core packages when invoked and also bundles numerous other packages that share a design philosophy, common grammar, and data structures
── Attaching core tidyverse packages ──────── tidyverse 2.0.0 ──
✔ dplyr 1.1.2 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.0
✔ ggplot2 3.4.2 ✔ tibble 3.2.1
✔ lubridate 1.9.2 ✔ tidyr 1.3.0
✔ purrr 1.0.1
── Conflicts ────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package to force all conflicts to become errors
tidyverse for data science
setup: penguins
# A tibble: 344 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm
<fct> <fct> <dbl> <dbl> <int>
1 Adelie Torgers… 39.1 18.7 181
2 Adelie Torgers… 39.5 17.4 186
3 Adelie Torgers… 40.3 18 195
4 Adelie Torgers… NA NA NA
5 Adelie Torgers… 36.7 19.3 193
6 Adelie Torgers… 39.3 20.6 190
# ℹ 338 more rows
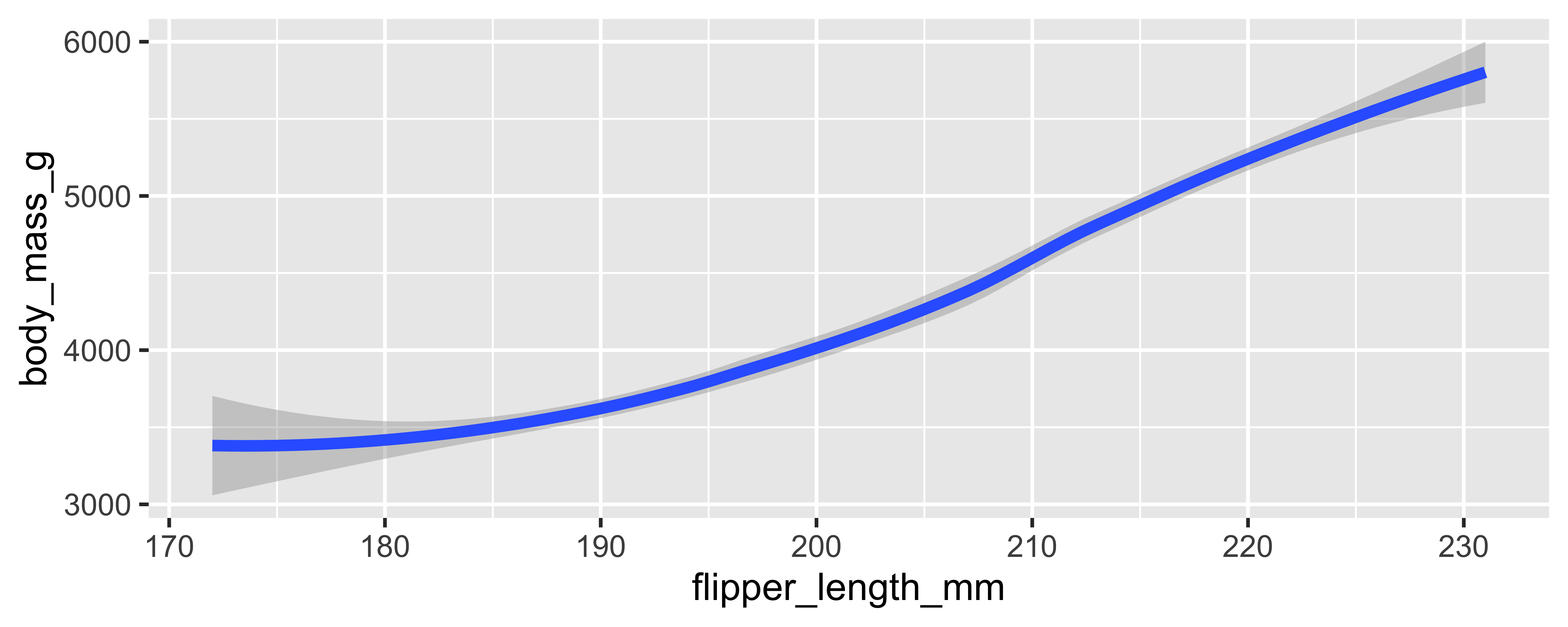
# ℹ 3 more variables: body_mass_g <int>, sex <fct>, year <int>a typical tidyverse pipeline
a typical tidyverse workflow
`summarise()` has grouped output by 'species'. You can override
using the `.groups` argument.# A tibble: 8 × 3
# Groups: species [3]
species sex mean_bw
<fct> <fct> <dbl>
1 Adelie female 3369.
2 Adelie male 4043.
3 Adelie <NA> NA
4 Chinstrap female 3527.
5 Chinstrap male 3939.
6 Gentoo female 4680.
# ℹ 2 more rowsa typical tidyverse workflow
penguins |>
drop_na(sex, body_mass_g) |>
group_by(species, sex) |>
summarize(mean_bw = mean(body_mass_g))`summarise()` has grouped output by 'species'. You can override
using the `.groups` argument.# A tibble: 6 × 3
# Groups: species [3]
species sex mean_bw
<fct> <fct> <dbl>
1 Adelie female 3369.
2 Adelie male 4043.
3 Chinstrap female 3527.
4 Chinstrap male 3939.
5 Gentoo female 4680.
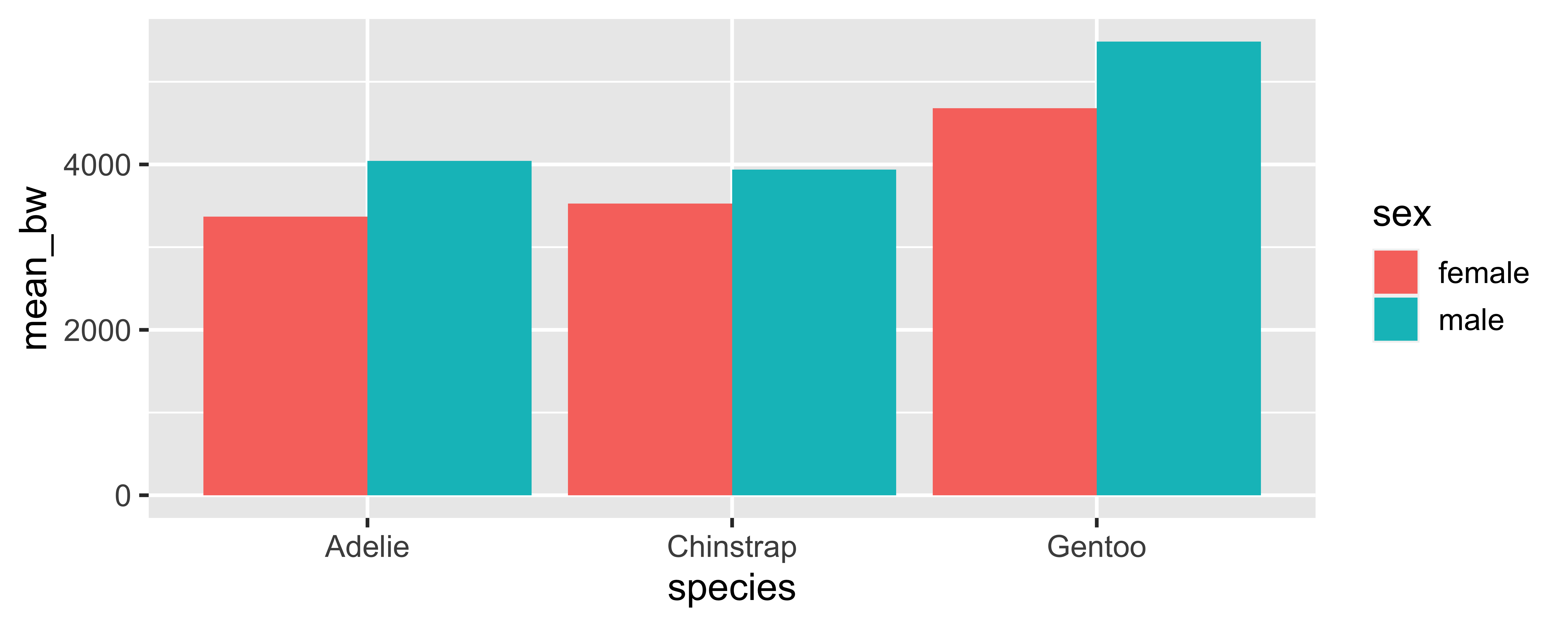
6 Gentoo male 5485.a typical tidyverse workflow
penguins |>
drop_na(sex, body_mass_g) |>
group_by(species, sex) |>
summarize(mean_bw = mean(body_mass_g), .groups = "drop")# A tibble: 6 × 3
species sex mean_bw
<fct> <fct> <dbl>
1 Adelie female 3369.
2 Adelie male 4043.
3 Chinstrap female 3527.
4 Chinstrap male 3939.
5 Gentoo female 4680.
6 Gentoo male 5485.a typical tidyverse workflow
a typical tidyverse workflow
a note about this presentation
- Sometimes I’ll show two options, where the now option is what you should do now.
- And sometimes I’ll show two options, where the now option is what you can do now.
- There will be more of the latter than the former!
- I’ll also sprinkle in some teaching tips along the way.
tidyverse 2.0.0
what’s new in tidyverse 2.0.0?
- lubridate is now a core tidyverse package
- package loading message advertises the conflicted package
lubridate - now core
lubridate, a package that makes it easier to do the things R does with date-times, is now a core tidyerse package.

lubridate - functionality
lubridate is most useful for parsing numbers or text that repsent dates into date-time:
Slightly more complex:
conflicted - now advertised
- Load tidyverse:
── Attaching core tidyverse packages ──────── tidyverse 2.0.0 ──
✔ dplyr 1.1.2 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.0
✔ ggplot2 3.4.2 ✔ tibble 3.2.1
✔ lubridate 1.9.2 ✔ tidyr 1.3.0
✔ purrr 1.0.1
── Conflicts ────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package to force all conflicts to become errors- Explicitly check for conflicts with
tidyverse::tidyverse_conflicts():
conflict resolution with base R
R’s default conflict resolution gives precedence to the most recently loaded package
- Before loading tidyverse - calling
filter()usesstats::filter():
Error in eval(expr, envir, enclos): object 'species' not found- After loading tidyverse - calling
filter()silently usesdplyr::filter():
# A tibble: 152 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm
<fct> <fct> <dbl> <dbl> <int>
1 Adelie Torgers… 39.1 18.7 181
2 Adelie Torgers… 39.5 17.4 186
3 Adelie Torgers… 40.3 18 195
# ℹ 149 more rows
# ℹ 3 more variables: body_mass_g <int>, sex <fct>, year <int>conflict resolution with conflicted
After loading conflicted - filter() doesn’t silently use dplyr::filter():
Error:
! [conflicted] filter found in 2 packages.
Either pick the one you want with `::`:
• dplyr::filter
• stats::filter
Or declare a preference with `conflicts_prefer()`:
• `conflicts_prefer(dplyr::filter)`
• `conflicts_prefer(stats::filter)`conflict resolution with conflicted - option 1
Pick the one you want with :::
# A tibble: 152 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm
<fct> <fct> <dbl> <dbl> <int>
1 Adelie Torgers… 39.1 18.7 181
2 Adelie Torgers… 39.5 17.4 186
3 Adelie Torgers… 40.3 18 195
# ℹ 149 more rows
# ℹ 3 more variables: body_mass_g <int>, sex <fct>, year <int>conflict resolution with conflicted - option 2
declare a preference with conflicts_prefer():
[conflicted] Will prefer dplyr::filter over any other package.# A tibble: 152 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm
<fct> <fct> <dbl> <dbl> <int>
1 Adelie Torgers… 39.1 18.7 181
2 Adelie Torgers… 39.5 17.4 186
3 Adelie Torgers… 40.3 18 195
# ℹ 149 more rows
# ℹ 3 more variables: body_mass_g <int>, sex <fct>, year <int>teaching tip
Don’t hide startup messages from teaching materials
Instead, address them early on to
- Encourage reading and understanding messages, warnings, and errors
- Help during hard-to-debug situations resulting from base R’s silent conflict resolution
But… Do teach students how to hide them in reports, particularly during editing/polishing stage!
dplyr 1.1.2
what’s new in dplyr 1.1.2?
A (non-exhaustive) list:
- Improved and expanded
_join()functionality - Added functionality for per operation grouping
- Quality of life improvements:
case_when()andif_else() - and more…
improved and expanded _join() functionality
- New
join_by()function for thebyargument in*_join()functions - Handling various matches (one-to-one, one-to-many, many-to-many relationships, etc.) and unmatched cases
- and more…
join_by()
setup: islands
We have the following information on the three islands we have penguins from:
join_by()
with by:
# A tibble: 344 × 3
species island coordinates
<fct> <chr> <chr>
1 Adelie Torgersen 64°46′S 64°5′W
2 Adelie Torgersen 64°46′S 64°5′W
3 Adelie Torgersen 64°46′S 64°5′W
4 Adelie Torgersen 64°46′S 64°5′W
5 Adelie Torgersen 64°46′S 64°5′W
6 Adelie Torgersen 64°46′S 64°5′W
# ℹ 338 more rowswith join_by():
penguins |>
left_join(
islands,
by = join_by(island == name)
) |>
select(species, island, coordinates)# A tibble: 344 × 3
species island coordinates
<fct> <chr> <chr>
1 Adelie Torgersen 64°46′S 64°5′W
2 Adelie Torgersen 64°46′S 64°5′W
3 Adelie Torgersen 64°46′S 64°5′W
4 Adelie Torgersen 64°46′S 64°5′W
5 Adelie Torgersen 64°46′S 64°5′W
6 Adelie Torgersen 64°46′S 64°5′W
# ℹ 338 more rowsteaching tip
Prefer join_by() over by
So that
- You can read it out loud as “where x is equal to y”, just like in other logical statements where
==is pronounced as “is equal to” - You don’t have to worry about
by = c(x = y)(which is invalid) vs.by = c(x = "y")(which is valid) vs.by = c("x" = "y")(which is also valid)
handling various matches
setup: three_penguins
Information about three penguins, one row per samp_id:
setup: weight_measurements
Information about weight measurements of these penguins, one row per samp_id, meas_id combination:
setup: flipper_measurements
Information about flipper length measurements of these penguins, one row per samp_id, meas_id combination:
one-to-many relationships - all good!
# A tibble: 6 × 5
samp_id species island meas_id body_mass_g
<dbl> <chr> <chr> <dbl> <dbl>
1 1 Adelie Torgersen 1 3220
2 1 Adelie Torgersen 2 3250
3 2 Gentoo Biscoe 1 4730
4 2 Gentoo Biscoe 2 4725
5 3 Chinstrap Dream 1 4000
6 3 Chinstrap Dream 2 4050many-to-many relationships - warning
What does the following warning mean?
Warning in left_join(weight_measurements, flipper_measurements, join_by(samp_id)): Detected an unexpected many-to-many relationship between `x` and
`y`.
ℹ Row 1 of `x` matches multiple rows in `y`.
ℹ Row 1 of `y` matches multiple rows in `x`.
ℹ If a many-to-many relationship is expected, set `relationship
= "many-to-many"` to silence this warning.# A tibble: 12 × 5
samp_id meas_id.x body_mass_g meas_id.y flipper_length_mm
<dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 3220 1 193
2 1 1 3220 2 195
3 1 2 3250 1 193
4 1 2 3250 2 195
5 2 1 4730 1 214
6 2 1 4730 2 216
7 2 2 4725 1 214
8 2 2 4725 2 216
9 3 1 4000 1 203
10 3 1 4000 2 203
11 3 2 4050 1 203
12 3 2 4050 2 203many-to-many relationships - explosion of rows
We followed the warning’s advice. Does the following look correct?
weight_measurements |>
left_join(flipper_measurements, join_by(samp_id), relationship = "many-to-many")# A tibble: 12 × 5
samp_id meas_id.x body_mass_g meas_id.y flipper_length_mm
<dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 3220 1 193
2 1 1 3220 2 195
3 1 2 3250 1 193
4 1 2 3250 2 195
5 2 1 4730 1 214
6 2 1 4730 2 216
7 2 2 4725 1 214
8 2 2 4725 2 216
9 3 1 4000 1 203
10 3 1 4000 2 203
11 3 2 4050 1 203
12 3 2 4050 2 203many-to-many relationships - rethink join_by()
setup: four_penguins
Information about three penguins, one row per samp_id:
# A tibble: 4 × 3
samp_id species island
<dbl> <chr> <chr>
1 1 Adelie Torgersen
2 2 Gentoo Biscoe
3 3 Chinstrap Dream
4 4 Adelie Biscoe unmatched rows - poof!
# A tibble: 6 × 5
samp_id meas_id body_mass_g species island
<dbl> <dbl> <dbl> <chr> <chr>
1 1 1 3220 Adelie Torgersen
2 1 2 3250 Adelie Torgersen
3 2 1 4730 Gentoo Biscoe
4 2 2 4725 Gentoo Biscoe
5 3 1 4000 Chinstrap Dream
6 3 2 4050 Chinstrap Dream unmatched rows - error
The unmatched argument protects you from accidentally dropping rows during a join:
unmatched rows - option 1
Use inner_join():
# A tibble: 6 × 5
samp_id meas_id body_mass_g species island
<dbl> <dbl> <dbl> <chr> <chr>
1 1 1 3220 Adelie Torgersen
2 1 2 3250 Adelie Torgersen
3 2 1 4730 Gentoo Biscoe
4 2 2 4725 Gentoo Biscoe
5 3 1 4000 Chinstrap Dream
6 3 2 4050 Chinstrap Dream unmatched rows - option 2
Set unmatched = "drop":
# A tibble: 6 × 5
samp_id meas_id body_mass_g species island
<dbl> <dbl> <dbl> <chr> <chr>
1 1 1 3220 Adelie Torgersen
2 1 2 3250 Adelie Torgersen
3 2 1 4730 Gentoo Biscoe
4 2 2 4725 Gentoo Biscoe
5 3 1 4000 Chinstrap Dream
6 3 2 4050 Chinstrap Dream unmatched rows - option 3
Do nothing – at your own risk!
# A tibble: 6 × 5
samp_id meas_id body_mass_g species island
<dbl> <dbl> <dbl> <chr> <chr>
1 1 1 3220 Adelie Torgersen
2 1 2 3250 Adelie Torgersen
3 2 1 4730 Gentoo Biscoe
4 2 2 4725 Gentoo Biscoe
5 3 1 4000 Chinstrap Dream
6 3 2 4050 Chinstrap Dream and more…
Inequality joins and rolling joins, made possible by join_by() being able to take expressions involving >, <=, etc.
Learn more about inequality joins at https://www.tidyverse.org/blog/2023/01/dplyr-1-1-0-joins/#inequality-joins
Learn more about rolling joins at https://www.tidyverse.org/blog/2023/01/dplyr-1-1-0-joins/#rolling-joins
What are inequality joins and rolling joins?
IYKYK!
If not, R4DS, 2nd Ed - Non-equi joins section is a great place to learn about them!
teaching tip
Exploding joins can be hard to debug for students!
Teach students how to diagnose whether the join they performed, and that may not have given an error, is indeed the one they wanted to perform. Did they lose any cases? Did they gain an unexpected amount of cases? Did they perform a join without thinking and take down the entire teaching server? These things happen, particularly if students are working with their own data for an open-ended project!
added functionality for per operation grouping
persistent grouping - handle with .groups
Remember our “typical tidyverse pipeline”? Why did we set .groups = "drop" in summarize()?
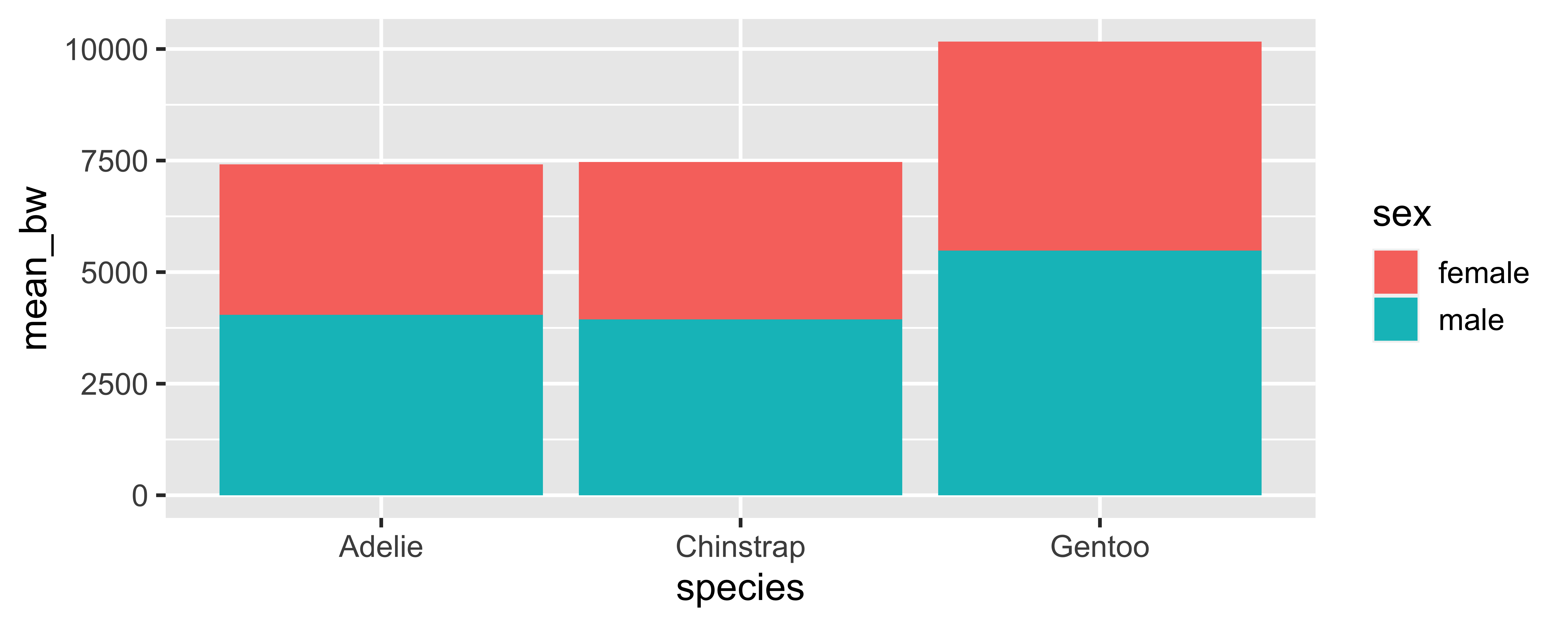
persistent grouping - message
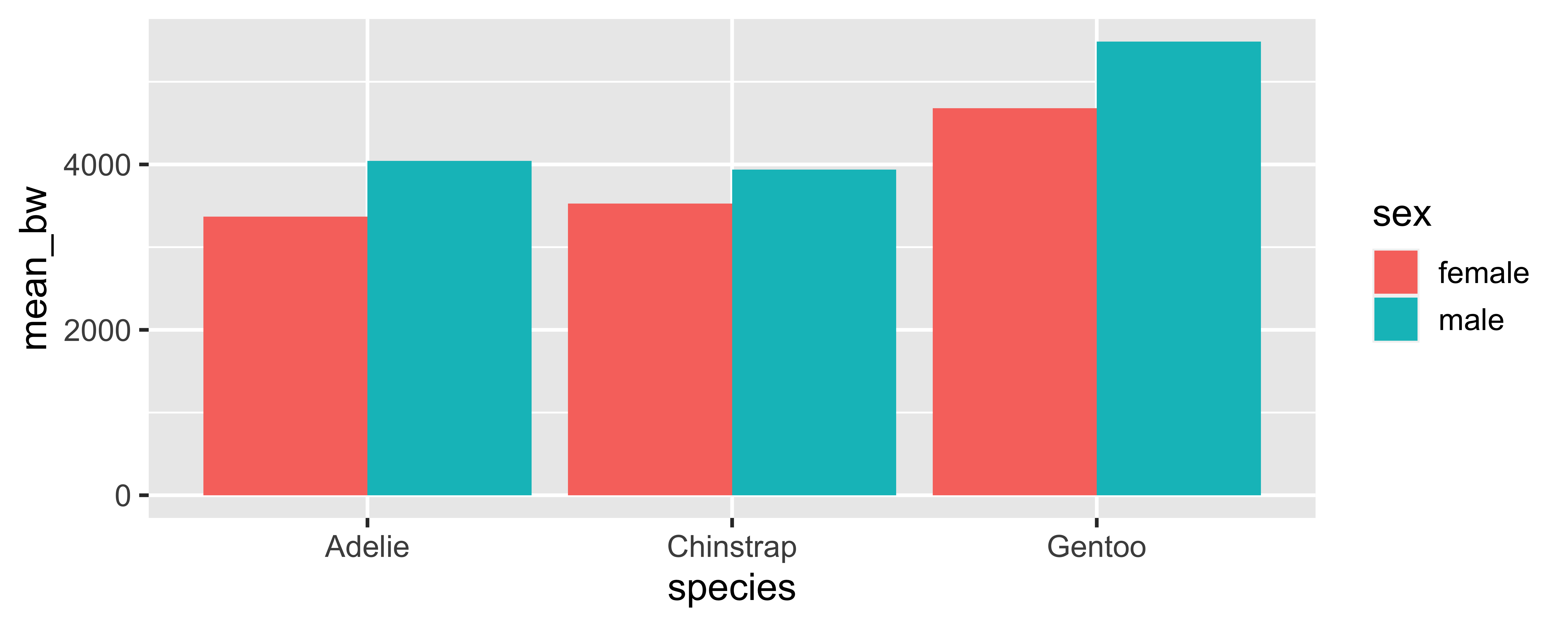
What if we don’t set it? Why does summarize() emit a message even though the result doesn’t change?
penguins |>
drop_na(sex, body_mass_g) |>
group_by(species, sex) |>
summarize(mean_bw = mean(body_mass_g)) |>
ggplot(aes(x = species, y = mean_bw, fill = sex)) +
geom_col(position = "dodge")`summarise()` has grouped output by 'species'. You can override
using the `.groups` argument.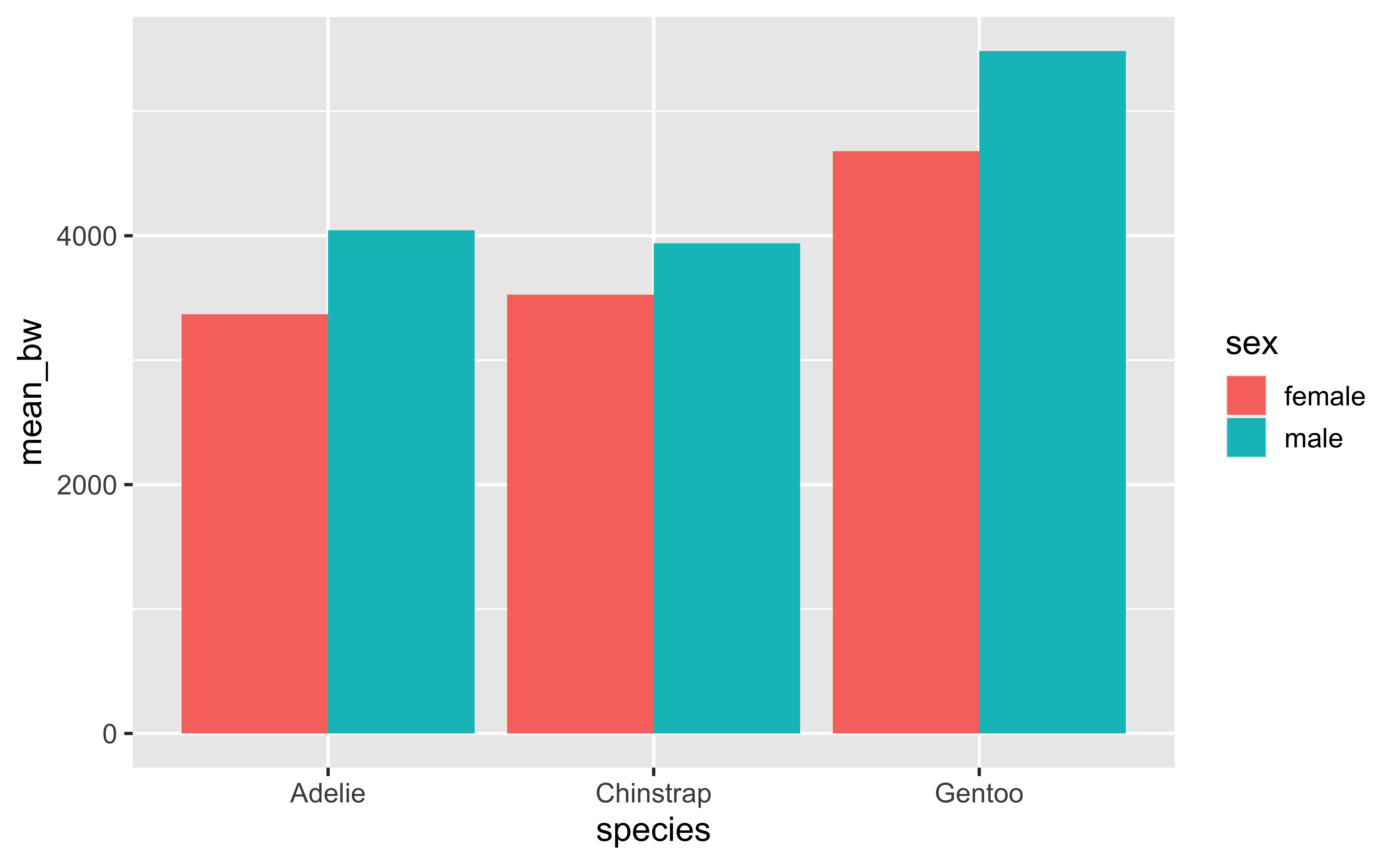
persistent grouping - downstream effects
persistent groups:
penguins |>
drop_na(sex, body_mass_g) |>
group_by(species, sex) |>
summarize(mean_bw = mean(body_mass_g)) |>
slice_head(n = 1)`summarise()` has grouped output by 'species'. You can override
using the `.groups` argument.# A tibble: 3 × 3
# Groups: species [3]
species sex mean_bw
<fct> <fct> <dbl>
1 Adelie female 3369.
2 Chinstrap female 3527.
3 Gentoo female 4680.persistent grouping - downstream effects
persistent groups:
penguins |>
drop_na(sex, body_mass_g) |>
group_by(species, sex) |>
summarize(mean_bw = mean(body_mass_g)) |>
gt::gt()`summarise()` has grouped output by 'species'. You can override
using the `.groups` argument.| sex | mean_bw |
|---|---|
| Adelie | |
| female | 3368.836 |
| male | 4043.493 |
| Chinstrap | |
| female | 3527.206 |
| male | 3938.971 |
| Gentoo | |
| female | 4679.741 |
| male | 5484.836 |
dropped groups:
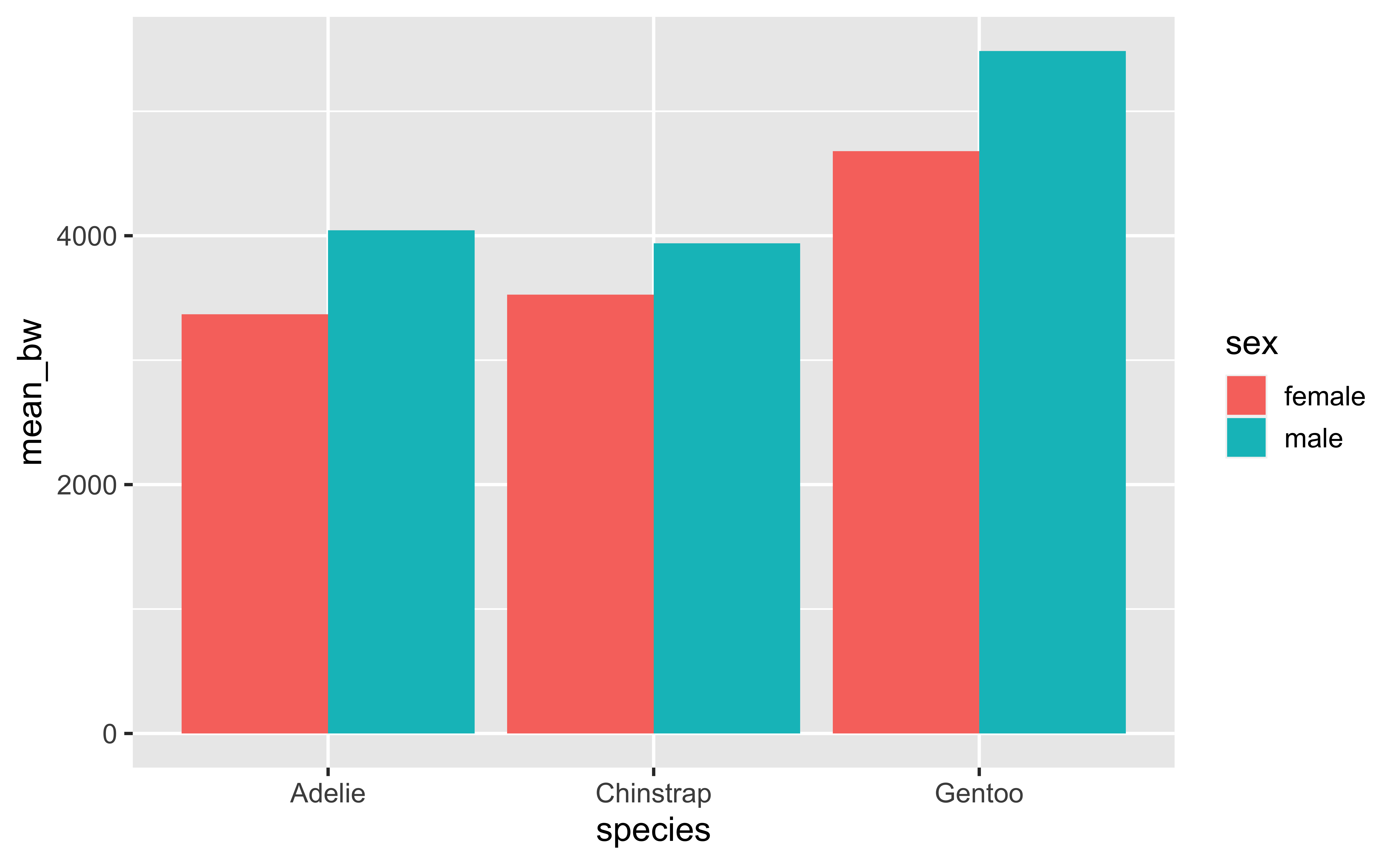
handling grouping - option 1
What we’ve already seen, explicitly selecting what to do with groups with .groups:
drop groups:
penguins |>
drop_na(sex, body_mass_g) |>
group_by(species, sex) |>
summarize(
mean_bw = mean(body_mass_g),
.groups = "drop"
)# A tibble: 6 × 3
species sex mean_bw
<fct> <fct> <dbl>
1 Adelie female 3369.
2 Adelie male 4043.
3 Chinstrap female 3527.
4 Chinstrap male 3939.
5 Gentoo female 4680.
6 Gentoo male 5485.keep groups:
penguins |>
drop_na(sex, body_mass_g) |>
group_by(species, sex) |>
summarize(
mean_bw = mean(body_mass_g),
.groups = "keep"
)# A tibble: 6 × 3
# Groups: species, sex [6]
species sex mean_bw
<fct> <fct> <dbl>
1 Adelie female 3369.
2 Adelie male 4043.
3 Chinstrap female 3527.
4 Chinstrap male 3939.
5 Gentoo female 4680.
6 Gentoo male 5485.handling grouping - option 2
Using per-operation grouping with .by:
group by 1 var:
group by 2+ vars:
group_by() vs. .by
group_by()is not superseded and not going away,.byis an alternative if you want per-operation grouping.Some verbs take
byinstead of.byas an argument 😞, but they come with informative errors 🙂.byalways returns an ungrouped data frameYou can’t create variables on the fly in
.by, you must create them earlier in your pipeline, e.g., unlikedf |> group_by(month = floor_date(date, "month")).bydoesn’t sort grouping keys,group_by()always sorts keys in ascending order, which affects the results of verbs likesummarise(), e.g., see the species orders in the two previous slides
teaching tip
Choose one grouping method and stick to it
It doesn’t matter whether you use group_by() (followed by .groups, where needed) or .by.
- For new learners, pick one and stick to it.
- For more experienced learners, particularly those learning to design their own functions and packages, it can be interesting to go through the differences and evolution.
quality of life improvements
for case_when() and if_else():
- all else denoted by
.defaultforcase_when() - less strict about value type for both
setup: penguin_quantiles
case_when()
penguins |>
mutate(
bm_cat = case_when(
is.na(body_mass_g) ~ NA,
body_mass_g < 3550 ~ "Small",
between(body_mass_g, 3550, 4750) ~ "Medium",
.default = "Large"
)
) |>
relocate(body_mass_g, bm_cat)# A tibble: 344 × 9
body_mass_g bm_cat species island bill_length_mm bill_depth_mm
<int> <chr> <fct> <fct> <dbl> <dbl>
1 3750 Medium Adelie Torger… 39.1 18.7
2 3800 Medium Adelie Torger… 39.5 17.4
3 3250 Small Adelie Torger… 40.3 18
4 NA <NA> Adelie Torger… NA NA
5 3450 Small Adelie Torger… 36.7 19.3
6 3650 Medium Adelie Torger… 39.3 20.6
# ℹ 338 more rows
# ℹ 3 more variables: flipper_length_mm <int>, sex <fct>,
# year <int>if_else()
penguins |>
mutate(
bm_unit = if_else(!is.na(body_mass_g), paste(body_mass_g, "g"), NA)
) |>
relocate(body_mass_g, bm_unit)# A tibble: 344 × 9
body_mass_g bm_unit species island bill_length_mm bill_depth_mm
<int> <chr> <fct> <fct> <dbl> <dbl>
1 3750 3750 g Adelie Torge… 39.1 18.7
2 3800 3800 g Adelie Torge… 39.5 17.4
3 3250 3250 g Adelie Torge… 40.3 18
4 NA <NA> Adelie Torge… NA NA
5 3450 3450 g Adelie Torge… 36.7 19.3
6 3650 3650 g Adelie Torge… 39.3 20.6
# ℹ 338 more rows
# ℹ 3 more variables: flipper_length_mm <int>, sex <fct>,
# year <int>teaching tip
It’s a blessing to not have to introduce NA_character_ and friends
Especially not having to introduce it as early as if_else() and case_when(). Cherish it!
Different types of NAs are a good topic for a course on R as a programming language, statistical computing, etc. but not necessary for an intro course.
and more…
- Further simplify your
case_when()statements withcase_match() - Selecting columns inside a function like
mutate()orsummarize()withpick() - Reproducibility and performance updates to
arrange() - Read more at https://www.tidyverse.org/tags/dplyr-1-1-0/
tidyr 1.3.0
new separate_*() functions
that supersede extract(), separate(), and separate_rows() because they have more consistent names and arguments, have better performance, and provide a new approach for handling problems:
| MAKE COLUMNS | MAKE ROWS | |
|---|---|---|
| Separate with delimiter | separate_wider_delim() |
separate_longer_delim() |
| Separate by position | separate_wider_position() |
separate_longer_position() |
| Separate with regular expression | separate_wider_regex() |
setup: three_penguin_descriptions
three_penguin_descriptions <- tribble(
~id, ~description,
1, "Species: Adelie, Island - Torgersen",
2, "Species: Gentoo, Island - Biscoe",
3, "Species: Chinstrap, Island - Dream",
)
three_penguin_descriptions# A tibble: 3 × 2
id description
<dbl> <chr>
1 1 Species: Adelie, Island - Torgersen
2 2 Species: Gentoo, Island - Biscoe
3 3 Species: Chinstrap, Island - Dream separate_wider_delim()
separate_wider_regex()
If you’re into that sort of thing…
enhanced reporting when things fail
previously
teaching tip
Excel text-to-column users are used to different approaches to “separate”
If teaching folks coming from doing data manipulation in spreadsheets, leverage that to motivate different types of separate_*() functions, and show the benefits of programming over point-and-click software for more advanced operations like separating longer and separating with regular expressions.
ggplot2 3.4.0
linewidth
previously
teaching tip
Check the output of your old teaching materials thoroughly
To not make a fool of yourself when teaching 🤣
and more…
other tidyverse updates
- Better stringr and tidyr alignment
- Ability to distinguish between
NAin levels vs.NAin values in forcats - New (more straightforward to learn/teach!) API for rvest
- Shorter, more readable, and (in some cases) faster SQL queries in dbplyr
- and more …
keeping up with the tidyverse
If you are interested in closely keeping up with updates in the tidyverse, the Tidyverse blog is the best place to read!
other tidyverse adjacent developments
The Tidyverse blog is also a great place to keep up with
tidymodels updates
and the magic that is webR! ✨
learn more
For a comprehensive overview of data science with R and the tidyverse, read the recently updated, and very soon to be available in paperback, R for Data Science, 2nd Edition.
thank you!
References
Packages
tidyverse: Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H (2019). “Welcome to the tidyverse.” Journal of Open Source Software, 4(43), 1686. doi:10.21105/joss.01686 https://doi.org/10.21105/joss.01686.
palmerpenguins: Horst AM, Hill AP, Gorman KB (2020). palmerpenguins: Palmer Archipelago (Antarctica) penguin data. R package version 0.1.0. https://allisonhorst.github.io/palmerpenguins/. doi: 10.5281/zenodo.3960218.
conflicted: Wickham H (2023). conflicted: An Alternative Conflict Resolution Strategy. R package version 1.2.0, https://CRAN.R-project.org/package=conflicted.
gt: Iannone R, Cheng J, Schloerke B, Hughes E, Lauer A, Seo J (2023). gt: Easily Create Presentation-Ready Display Tables. R package version 0.9.0, https://CRAN.R-project.org/package=gt.